Publications
Highlights
(For a full list see below or go to Google Scholar)
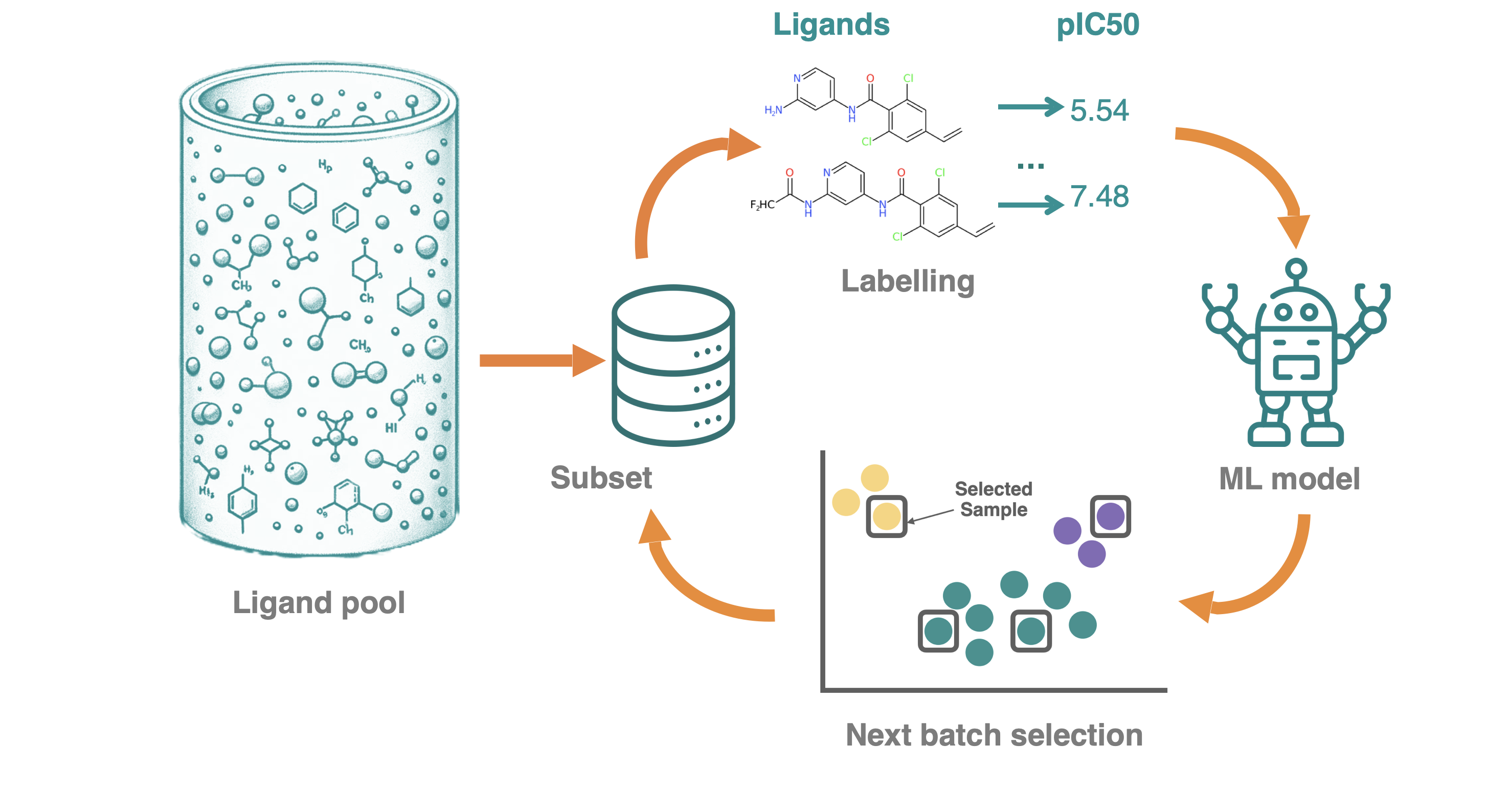
Benchmarking active learning trategies for best performance on labelling data for binding affinities
R. Gorantla, A. Kubincova, B. Suutari, B. P. Cossins, A. S.J.S. Mey
The paper was featured in Practical Cheminformatics
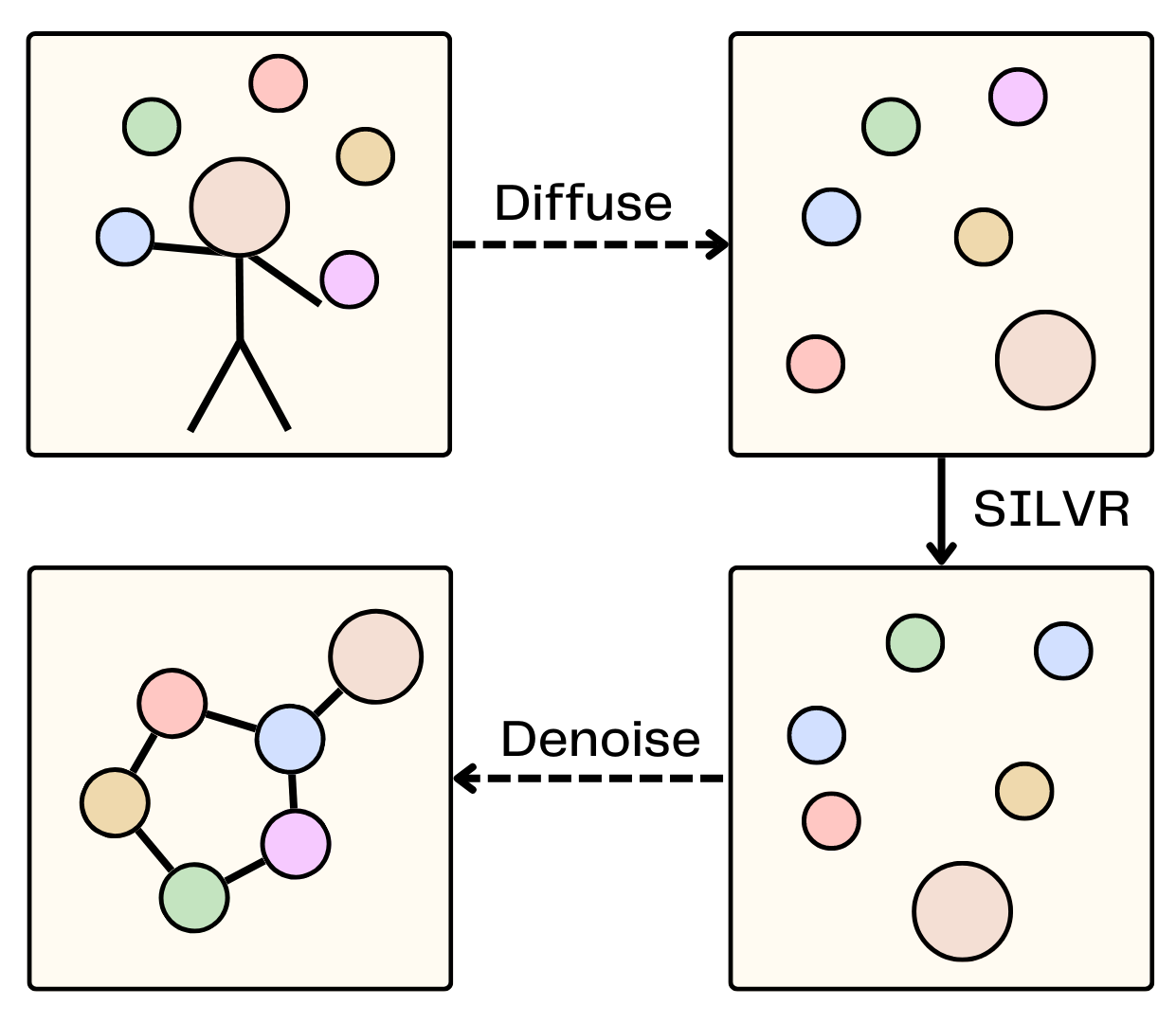
SILVR is a method for conditioning a pre-trained equivariant diffusion model to generate new molecules from X-ray fragements.
Nicholas T. Runcie, Antonia S.J.S. Mey
J. Chem. Inf. Model. 63, 19, 5996–6005 (2023)
The paper was selected as an Editor’s Choice article
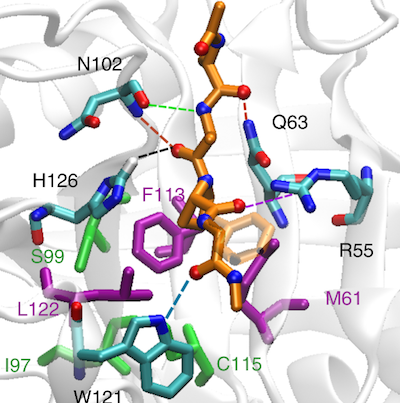
Molecular simulations can give an alternative explanation for the reduced catalytic rate of different Cyclophilin A mutants that is experimentally testable.
P. Wapeesittipan, A.S.J.S. Mey, M. Walkinshaw, J. Michel
Accounting for differently charged ligands can be difficult in simulations, a framework for charging corrections was tested as part of a blinded challenge.
A.S.J.S. Mey, J. Juárez-Jiménez, J. Michel
J. Comput. Aided. Mol. Des. 32, 199 (2018)
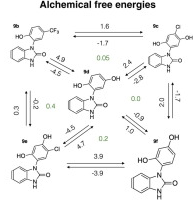
Alchemical free energy calculation for HSP90-α using SOMD give competitive results in a blinded prediction challenge.
A.S.J.S. Mey, J. Juárez-Jiménez, A. Hennessy, J. Michel
Bioorg. Med. Chem. 24, 4890 (2016)
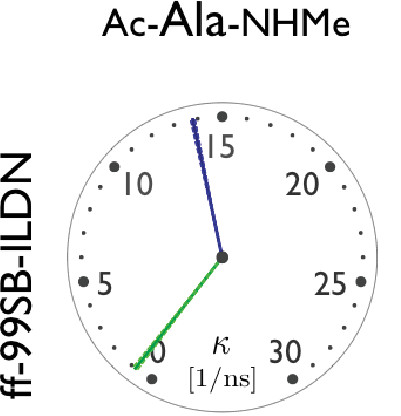
Protein dynamics is highly dependent on the choice of molecular forcefield. The speed of different forcefields is assessed.
F. Vitalini, A.S.J.S. Mey, F. Noé and B.G. Keller
J. Chem. Phys. 142, 084101 (2015)
Most read article of in JCP in 2015
Preprints
- Protocols for metallo- and serine-β-lactamase free energy predictions: insights from cross-class inhibitors J. J. Güven, M. Hanževački, P. Kalita, A. J.Mulholland, A. S.J.S. Mey
- Comparison of Methodologies for Absolute Binding Free Energy Calculations of Ligands to Intrinsically Disordered Proteins M. Papadourakis, Z. Cournia, A. S.J.S. Mey, J. Michel
- mRNA interactions with disordered regions control protein activity Y. Luo, S. Pratihar, E. Horste, S. Mitschka, A. S.J.S. Mey, H.M. Al-Hashimi, C. Mayr
chemRxiv 2024-9ksxr
bioRxiv 2024.07.19.604182
bioRxiv 2023.02.18.529068
Full List
- Digital skills in chemical education A. R. McCluskey, M. Rivera, A. S.J.S. Mey
- Dirac--Bianconi Graph Neural Networks -- Enabling Non-Diffusive Long-Range Graph Predictions C. Nauck, R. Gorantla, M. Lindner, K. Schürholt, A. S.J.S. Mey, F. Hellmann
- Sire: An Interoperability Engine for Prototyping Algorithms and Exchanging Information Between Molecular Simulation Programs C. Woods, L. Hedges, A. J. Mulholland, M. Malaisree, P. Tosco, H. Loeffler, M. Suruzhon, M. Burman, S. Bariami, S. Bosisio, G. Calabro, F. Clark, A. S.J.S. Mey, J. Michel
- Benchmarking active learning protocols for ligand binding affinity prediction R. Gorantla, A. Kubincova, B. Suutari, B. P. Cossins, A. S.J.S. Mey
- Markov State Models: to optimize or not to optimize R. Arbon, Y. Zhu, A. S.J.S. Mey
- A suite of tutorials for the BioSimSpace framework for interoperable biomolecular simulation L. O. Hedges, S. Bariami, M. Burman, F. Clark, B. P. Cossins, A. Hardie, A. M. Herz, D. Lukauskis, A. S.J.S. Mey, J. Michel, J. Scheen, M. Suruzhon, C. J. Woods, Z. Wu
- From Proteins to Ligands: Decoding Deep Learning Methods for Binding Affinity Prediction Rohan Gorantla, Alzbeta Kubincova, Andrea Y. Weisse, Antonia S.J.S. Mey
- SILVR: Guided Diffusion for Molecule Generation Nicholas T. Runcie, Antonia S.J.S. Mey
- Course Materials for an Introduction to Data-Driven Chemistry James Cumby, Valentina Erastova, Matteo T. Degiacomi, J. Jasmin Güven, Claire L. Hobday, Antonia S.J.S. Mey, Hannah Pollak, Rafal Szabla
- What geometrically constrained models can tell us about real-world protein contact maps J. Jasmin Güven, Nora Molkenthin, Steffen Mühle, Antonia S.J.S. Mey
- Efficient Purification of Cowpea Chlorotic Mottle Virus by a Novel Peptide Aptamer G. Tscheuschner, M. Ponader, C. Raab, P. S. Weider, R. Hartfiel, J. O. Kaufmann, J. L. Völzke, G. Bosc-Bierne, C. Prinz, T. Schwaar, P. Andrle, H. Bäßler, K. Nguyen, Y. Zhu, A.S.J.S. Mey, A. Mostafa, I. Bald, M. G. Weller
- Chapter 8: The IMAGINARY Journey to Open Mathematics Engagement: History and Current Projects E. Londaits, A. Matt, A.S.J.S. Mey, D. Ramos, C. Stussak, B. Violet
- Best practices for constructing, preparing, and evaluating protein-ligand binding affinity benchmarks D.F. Hahn, C.I. Bayly, H.E. Bruce Macdonald, J.D. Chodera, A.S.J.S. Mey, D.L. Mobley, L. Perez Benito, C.E. M. Schindler, G.Tresadern, G.L. Warren
- Dynamic Profiling of β-Coronavirus 3CL Mpro Protease Ligand-Binding Sites E. Cho, M. Rosa, R. Anjum, S. Mehmood, M. Soban, M. Mujtaba, K. Bux, S. C. Dantu, A. Pandini, J. Yin, H. Ma, A. Ramanathan, B. Islam, A.S.J.S. Mey, D. Bhowmik, S. Haider
- Implementation of the QUBE Force Field in SOMD for High-Throughput Alchemical Free-Energy Calculations L. Nelson, S. Bariami, C. Ringrose, J. Horton, V. Kurdekar, A.S.J.S. Mey, J. Michel, D. Cole
- Best Practices for Alchemical Free Energy Calculations A.S.J.S. Mey, B. Allen, H.E. Bruce Macdonald, J.D. Chodera, D. Hahn, M. Kuhn, J. Michel, D.L. Mobley, L.N. Naden, S. Prasad, A. Rizzi, J. Scheen, M.R. Shirts, G. Tresadern, H. Xu
- A Hybrid Alchemical Free Energy and Machine Learning Methodology for the Calculation of Absolute Hydration Free Energies of Small Molecules J. Scheen, W. Wu, A.S.J.S. Mey, P. Tosco, M. Mackey, J. Michel
- Assessment of Binding Affinity via Alchemical Free-Energy Calculations M. Kuhn, S. Firth-Clark, P. Tosco, A.S.J.S. Mey, M. Mackey, J. Michel
- Self-organized emergence of folded protein-like network structures from geometric constraints N. Molkenthin, S. Mühle, A.S.J.S. Mey, M. Timme
- Dynamic design: manipulation of millisecond timescale motions on the energy landscape of Cyclophilin A J. Juárez-Jiménez, A. Gupta, G. Karunanithy, A.S.J.S. Mey, et al.
- BioSimSpace: An interoperable Python framework for biomolecular simulation L.O. Hedges, A.S.J.S. Mey, C.A. Laughton, et al.
- Allosteric effects in a catalytically impaired variant of the enzyme Cyclophilin A may be explained by changes in nano-microsecond time scale motions P. Wapeesittipan, A.S.J.S. Mey, M. Walkinshaw, J. Michel
- Effect of automation on the accuracy of alchemical free energy calculation protocols over a set of ACK1 inhibitors J.M. Granadino-Roldan*, A.S.J.S. Mey*, J.J. Perez, S. Bosisio, J. Rubio-Martinez, J. Michel
- Impact of domain knowledge on blinded predictions of binding energies by alchemical free energy calculations A.S.J.S. Mey, J. Juárez-Jiménez, J. Michel
- Blinded predictions of host-guest standard free energies of binding in the SAMPL5 challenge S. Bosisio, A.S.J.S. Mey, J. Michel,
- Analytical methods for structural ensembles and dynamics of intrinsically disordered proteins M. Schor, A.S.J.S. Mey, C.E. MacPhee
- Blinded predictions of distribution coefficients in the SAMPL5 challenge S. Bosisio, A.S.J.S. Mey, J. Michel
- Blinded predictions of binding modes and energies of HSP90-α ligands for the 2015 D3R Grand Challenge A.S.J.S. Mey*, J. Juárez-Jiménez*, A. Hennessy, J. Michel
- Elucidation of Non-Additive Effects in Protein-Ligand Binding Energies: Thrombin as a Case Study, G. Calabrò, C.J. Woods, F. Powlesland, A.S.J.S Mey, A.J. Mulholland, J. Michel
- Shedding light on the dock-lock mechanism in amyloid fibril growth using Markov State Models M. Schor, A.S.J.S. Mey, F. Noé, C.E. MacPhee
- Dynamic Properties of Forcefields F. Vitalini*, A.S.J.S. Mey*, F. Noé and B.G. Keller
- Statistically optimal analysis of state-discretized trajectory data from multiple thermodynamic states H. Wu, A.S.J.S. Mey, E. Rosta, F. Noé
- xTRAM: Estimating equilibrium expectations from time-correlated simulation data at multiple thermodynamic states A.S.J.S. Mey, H. Wu, and F. Noé
- Rare-event trajectory ensemble analysis reveals metastable dynamical phases in lattice proteins A.S.J.S. Mey, P.L. Geissler and J.P. Garrahan
- Variational approach to molecular kinetics F. Nüske, B.G. Keller, G. Pérez-Hernández, A.S.J.S. Mey, F. Noé
- EMMA - A software package for Markov model building and analysis M. Senne, B. Trendelkamp-Schroer, A.S.J.S. Mey, C. Schütte, F. Noé
- Thermodynamics of trajectories of the one-dimensional Ising model E.S. Loscar, A.S.J.S. Mey, J.P. Garrahan
Nat. Chem.
ICML workshop paper
J. Chem. Phys. 160, 202503 (2024)
J. Chem. Inf. Model. (2024)
J. Chem. Theory Comput. 20, 977–988 (2024)
Living Journal of Computational Molecular Science 5 (1), 2375-2375 (2023)
J. Chem. Inf. Model. (2023)
J. Chem. Inf. Model. 63, 19, 5996–6005 (2023)
J. Open Source Ed. 6, 192 (2023)
Phys. Biol. 20 046004 (2023)
Viruses, 15(3), 697 (2023)
World Scientific Series on Science Communication pp. 135-163 (2023)
Living J. Comp. Mol. Sci. 4(1), 1497 (2022)
J. Chem. Inf. Model. 61, 6, 3058–3073 (2021)
J. Chem. Inf. Model. 61, 5, 2124–2130 (2021)
Living J. Comp. Mol. Sci. 2 (1), 18378 (2020)
J. Chem. Inf. Model. 60, 11, 5331–5339 (2020)
J. Chem. Inf. Model. 60, 6, 3120–3130 (2020)
PLoS ONE 15(2), e0229230 (2020)
Chem. Sci., 11, 2670-2680 (2020)
JOSS, 4, 1831 (2019)
Comms. Chem. 2 41 (2019)
PloS One 14, e0213217 (2019)
J. Comput. Aided. Mol. Des. 32, 199 (2018)
J. Comput. Aided. Mol. Des. 31, 61 (2017)
Biophys. Rev. 8, 429 (2016)
J. Comput. Aided. Mol. Des. 30, 1101 (2016)
Bioorg. Med. Chem. 24, 4890 (2016)
J. Phys. Chem. B 120, 5340 (2016)
J. Phys. Chem. Lett. 6, 1076 (2015)
J. Chem. Phys. 142, 084101 (2015)
J. Chem. Phys. 141, 214106 (2014)
Phys. Rev. X 4, 041018 (2014)
Phys. Rev. E 89, 032109 (2014)
J. Chem. Theory Comput. 10, 1739 (2014)
J. Chem. Theory Comput. 8, 2223 (2012)
J. Stat. Mech. 2011, P12011 (2011)


